by Lynn Langit and Kelly Kermode
When building data analysis pipelines for genomic-scale workloads, the typical approach is to use a Data Lake Architecture. In this article, we’ll detail using this architecture on Azure. The general Data Lake pattern (shown below) includes several surface areas and uses common cloud services. Of note are the three major surface areas — Blob Storage for files (data layer), Batch Compute for Virtual Machine clusters (compute layer), and Interactive Analysis for query-on-files (analysis layers).
Files are the Data
In the Data Lake pattern, data, which is usually in the form of various types of files, is stored in BLOB storage containers. The compute plane in this pattern takes the form of a job controller which spawns a burstable cluster. Also, implementation of appropriate security via authentication, encryption, and endpoint protection is a key part of any solution.
Additionally, it is becoming increasingly common to add an optimization layer over BLOB storage. This is most commonly done using a FUSE library, which results in improved usability. Another recent pattern, which is particularly relevant for bioinformatics, is the incorporation of cloud-hosted genomic reference data into cloud pipelines.
Because the ever-increasing massive data volumes processed by the pipelines are in the form of files, building performant pipelines requires careful attention to the format and size of these input files. In genomics, there are a number of proprietary file types, such as FASTA, FASTQ, BAM, UBAM, SAM, VCF, and more. In conjunction with selecting and using the appropriate file type for the particular compute process (i.e. certain processes require certain input file types), pre-processing files (converting types, partitioning, etc…) is important in building optimized pipelines.
Genomic-scale data volumes are unprecedented.
Compute Choices
Another area of complexity in building genomic-scale pipelines is selecting the best-fit compute pattern. Most modern pipelines implement clusters of Docker container instances running on Virtual Machines (VMs). There are a number of key choices in building these clusters, starting with the size and type of the underlying VMs.
Note: careful attention to building efficient Docker container images is critical.
The type of job scheduler and controller used can vary. My team has built fast, scalable pipelines using the most common big data pipeline pattern: a Kubernetes controller and Apache Spark for in-memory distributed processing. It’s notable, that in some verticals, such as genomics, there are a number of alternative architectures.
Azure pipelines can be built with common Data Lake patterns such as those which use Azure Kubernetes Services and Docker containers — or — managed Apache Spark with Databricks or HDInsight. Microsoft Genomics has also built a domain-specific set of tools for cloud-scaling genomics which includes Microsoft Genomics Service (for secondary genomic analysis as shown below) and Cromwell-on-Azure.
Cromwell on Azure for Genomics
In bioinformatics, there is increasing collaboration between research labs globally on building reusable, scalable analysis tools. One organization, The Broad Institute at MIT and Harvard, has created a set of optimized tools and libraries, including the GATK toolkit and also an associated scalable workflow manager (Cromwell) and scripting language (WDL).
WDL stands for Workflow Definition Language; a WDL (‘widdle’) file indicates what actions to take with which input data files.
Cromwell is the workflow engine that coordinates the job executions launched by the trigger files for the WDL workflow tasks. Cromwell manages job executions defined by WDL on a number of configurable backend environments, including HPC, public cloud and others.
To test a reference Data Lake for genomic-scale analysis on Azure, we tried out the open-source solution on the CromwellOnAzure GitHub repo. This solution includes sample data and WDL files. It implements the Data Lake pattern using Azure services and components as shown below.
Setup the sample
If you want to try out this example yourself, plan for about one hour for the setup process. The actual setup time (script run) took 27 minutes for us on the `East US` Azure data center. Below are a couple of tips for setting up this sample yourself:
1. Create a dedicated Azure Resource Group for your test. This makes it easier to locate your resources and also to delete them when you are done testing.
2. Verify your quota allocation for low priority CPU cores for VMs. The example WDL scripts use between 1 to 16 low priority cores. New Azure accounts have quota set for this type of core to 0, so you would need to submit a quote increase request to be able to run the samples as configured. It can take a couple of days to get this quota approved, so do this step first.
NOTE: you may need to set up a ‘pay-as-you-go’ (rather than free tier) Azure account to get sufficient CPU quota to run the example.
3. Use the default service name prefix of ‘coa…..’ so that you can easily locate services implemented when you run the setup scripts.
4. Verify that you have the appropriate Azure role assignment (permission) on your Azure subscription as described in the `README.md` file in the repo.
An example of the Azure service instances that the sample deployment creates (after running the downloadable deployment executable file) in the ‘hello-azure’ Resource group is shown below.
NOTE: If you’d prefer to watch a short screencast (rather than setting up the sample yourself) which shows the sample running a cromwell job AFTER the setup is complete, then see the video linked below.
https://www.youtube.com/watch?v=yAeEmbP4RCc
Files are the Data
In this sample, the Data Lake pattern is implemented using a set of Azure Blob containers which contain named folders. By default, it includes the following Blob containers — cromwell-executions, cromwell-workflow-logs, inputs, outputs, and workflows. Note: folders are logical structures within Azure Blob containers.
To start a job execution, upload a `trigger.json` file to the `workflows\new` folder in the blob container.
This creates a JSON file in a working folder location named ‘workflows\inprogress` while the job is being run. After the job completes more metadata files are written & the ‘workflows\inprogress` folder & file is removed.
Tip: Refresh the BLOB container view frequently to see job execution progress updates. Also there are logs generated.
Job execution results are found in the cromwell-executions container in the associated job name and job run (GUID) and named result location, i.e. stdout, etc…
All of the examples use very small datasets, the sample does not demonstrate any other type of optimization of input files in pipelining (by converting file types, partitioning files, etc…).
Compute Choices
The sample creates a VM which includes Docker Compose with four Docker containers. This VM implements an instance of the Cromwell service in server mode (also called a cromwell server). Server mode starts Cromwell as a web server that exposes REST endpoints. This VM serves as the WDL job coordinator service.
The VM also hosts a C# service named `TriggerService.` The trigger service fires when a file is uploaded to a defined blob container folder location. The job controller VM hosts four Docker containers which run Cromwell, MySQL, TES or TriggerService instances.
The Azure Storage account is mounted to the cromwell server VM using blobfuse, which enables Azure Block Blobs to be mounted as a local file system available to all of the Docker containers running on the VM. An OS and data disk, network interface, public IP address, virtual network, and network security group are also created by the sample deployment utility.
A Cromwell server can be configured to use a number of backends on which to execute the distributed workflow tasks. This sample implements a version of the TES (Task Execution Service) backend for Cromwell. The TES API defines a GAG4H (genomics) standardized schema and API for describing batch execution tasks. A task defines a set of input files, a set of containers and commands to run, a set of output files, and some other logging and metadata.
To build dynamic the compute clusters, the sample uses the Azure Batch API as a downstream compute cluster generator. Azure Batch creates and manages a pool of compute nodes (VMs), installs applications, and schedules jobs to run on the nodes. Use Batch APIs and tools, command-line scripts, or the Azure portal to configure, manage, and monitor analysis jobs.
Use the Azure Batch API to run large-scale parallel batch jobs efficiently in Azure.
Microsoft wrote an open-source library (TES-Azure) which enables the Azure Batch API to be able to spawn compute clusters when invoked by a TES-compliant job. This cromwell server is configured to use the TES-Azure backend API which bundles the TES API and the Azure Batch API together. TES-Azure includes a number of key features:
Authentication options — standalone operation — or — multiple user auth (via AAD+OAuth) with per-tenant or per-user task isolation
File transfer for task inputs & outputs — reads from Azure Blob, HTTP, HTTPS, SFTP, FTP+SSL, AWS S3, GCP & writes to Azure Blob, SFTP, FTP+SSL
Security and Logging — secrets in Azure Key Vault & logging/tracing sent to Azure App Insights
Provisioning options for backend resources — through a REST API — or — via Docker-based deployments with Terraform
Container-based task execution from DockerHub — or — from a private container registry
Resource configuration for task executions — CPU, Memory and Disk
Note: the use of ‘low priority CPU cores’ (also called ‘low priority VMs’ in the Azure documentation) in the generated VM cluster — this is an important optimization which will save considerable money in scaled analysis pipelines.
Running a Workflow
Example WDL scripts specify the workflow name, task execution order, task definition, and task runtimes. Task runtimes normally execute in custom Docker container instances. Azure Batch coordinates each VM (set of cores) spawns Docker instances which execute the defined tasks in a fully distributed manner.
To kick off a job (or workflow), the researcher interacts with the solution using the Azure Blobstore. They will write files (or use the samples) of the following types:
WDL — WDL script file for cromwell workflow which describes tasks and workflows — example WDL file linked here
Inputs — JSON file which provides values for variables declared in WDL file, commonly includes paths to input genomic data files (FASTA, FASTQ, BAM, VCF…)
Trigger — JSON file which provides values for WDL and JSON file paths, commonly BLOB containers. An example trigger.json file shown below.
Using the simplest way to start the a workflow, the researcher uploads the trigger file to the specified blob folder location (`workflows\new\`) using the Azure portal. The TriggerService generates a job execution on trigger (file) upload. This job request is sent to the cromwell server.
The server then invokes the TES/Azure Batch API backend which in-turn spawns a dynamic cluster of cores (VMs) on which the job tasks will be run. A number of logs are generated during this process.
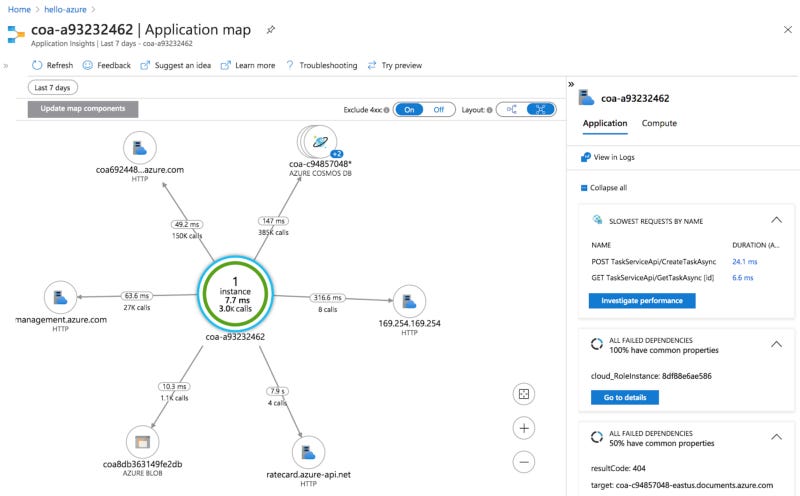
The researcher then views the status of an in-progress job in the folder `workflows\inprogress\nnnnnn….json`, noting the file name and the id number assigned to the job execution. The researcher can also view job task information using the CosmosDB NoSQL database using the SQL API for CosmosDB. They can monitor the job progress using the Azure Batch WebUI and or Azure Application Insights.
After the job execution completes, then the output from the job is put into the `cromwell-executions\folder..\…` path as described in the WDL file.
Monitoring
Azure Batch resource usage can be monitored using the console (shown below) or with other tools and logs. Additional included services are Application Insights (contains logs from TES and the Trigger Service to enable debugging) and the previously mentioned Cosmos DB (includes information and metadata about each TES task that is run as part of a workflow).
Next Steps: Verifying Scalability
While we were able to test the sample successfully using a number of linked WDL scripts, as noted previously, all of these examples use very small-sized input files. Given that, we consider our testing of this architecture not yet complete.
The goal of this pattern is to be able to efficiently process genomic-sized data pipelines. The next step in our work will be to test this example architecture with increasingly large input data files to get answers to questions such as the following:
What is the appropriate cluster size in terms of requested CPUs, VMs, VM configuration, etc… for real-world pipelines?
How does the caching mechanism affect job retries for (subsequent, stuck or failed) job runs?
Are there recommended optimizations for pre-processing files for known analysis job types, i.e. single cell RNA analysis, etc…?
As we continue this sample to test at scale, it will be interesting to further explore the monitoring available via Azure Application Insights, (one example system view is shown below).
Conclusions
As mentioned, it’s important to consider, test and compare the two general approaches to building Data Lake solutions for this genomics use case.
Kubernetes with containers — OR —
Domain-specific, vendor-specific cluster managers, such as cromwell
My team has had success at scale with the first approach (see our work with CSIRO for VariantSpark on AWS). Many large research organizations, such as The Broad Institute, favor the second approach (cromwell/WDL + Azure Batch) or AWS Batch or GCP Life Sciences/PAPI API).
In our ongoing work with bioinformatics clients, we’ll continue to report our results of testing cloud genomic pipelines patterns at scale. It is interesting to note that there is a GA4GH library named TESK which is an implementation of the TES interface that uses Kubernetes.
Links and References
Cromwell-on-Azure libraries https://github.com/microsoft/CromwellOnAzure
Learning Cromwell-on-Azure companion GitHub Repo
https://github.com/lynnlangit/learning-cromwell-on-azureTES and TES-Azure
https://github.com/ga4gh/task-execution-schemas & - https://github.com/microsoft/tes-azureTES back-end for cromwell
https://cromwell.readthedocs.io/en/develop/backends/TES/Genomics on Azure FAQ
https://docs.microsoft.com/en-us/azure/genomics/frequently-asked-questions-genomicsAzure Genomics Microsoft Reactor presentation, July 2020 (60 min)